CT radiomics, pathology, and genomics data can enable an artificial intelligence (AI) algorithm to predict if a non-small cell lung cancer (NSCLC) patient will respond to immunotherapy, according to research published August 29 in Nature Cancer.
A team of researchers from Memorial Sloan Kettering Cancer Center (MSKCC) developed a deep-learning model that integrates CT radiomics with histopathologic and genomic features to predict response to immunotherapy. In testing, the group found that the algorithm yielded an area under the curve (AUC) of 0.80, outperforming each individual feature alone.
"We show that existing data from multiple cancer diagnostic modalities can be annotated, abstracted and combined using computational and machine-learning methods for next-generation biomarker development in NSCLC immunotherapy response prediction," wrote co-first authors Rami Vanguri, PhD; Dr. Jia Luo; Andrew Aukerman; Jacklynn Egger; and colleagues.
Although immunotherapy is used to treat almost all patients with advanced NSCLC, the identification of robust predictive biomarkers remains a challenge, according to the researchers. To help address this ongoing issue, the MSKCC researchers sought to utilize AI and with clinical data to predict immunotherapy response.
The researchers gathered a cohort of 247 patients with advanced NSCLC who had received programmed death ligand-1 (PD-L1)-blockade-based therapy at MSKCC between 2014 and 2019. Of these, only 25% responded to immunotherapy.
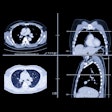
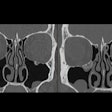
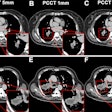
Next, they collected baseline data obtained for each patient during diagnostic clinical workup, including clinical information; contrast-enhanced chest CT scans; digitized PD-L1 immunohistochemistry slides of tissue containing NSCLC; genomic features from the MSK-IMPACT clinical next-generation sequencing platform; and their immunotherapy outcomes.
The clinical data were annotated by domain experts and then a computational workflow was developed to extract patient features. For the CT exams, radiomics features were extracted separately for lung parenchymal, pleural, and nodal lesions.
The researchers first quantified the individual predictive capacities for each modality. On its own, the CT-derived features produced only modest predictive performance.
The researchers then assembled a biomarker that was used to train and test a deep-learning model for risk prediction. The algorithm, called dynamic deep attention-based multiple-instance learning (DyAM), generates a partial risk score attributed to each feature, as well as a final score and the attention weight and share for each feature.
DyAM was validated using 10-fold cross-validation, and it achieved better results than the individual modalities for predicting immunotherapy response.
| AI model yields best immunotherapy response predictions | ||||
| Tumor mutational burden | CT radiomics features | PDL-1 immunohistochemistry score | All features combined | |
| AUC | 0.61 | 0.64 | 0.73 | 0.80 |
The results serve as a proof of principle for the value of integrating multiple features, according to the researchers.
"In the future, assembly of large well-annotated multi-institutional training datasets will lead to development of robust multimodal classifiers that serve as powerful biomarkers," the authors wrote. "These decision aids could be integrated into routine clinical care and used to quickly and precisely distinguish responders and nonresponders."