Large language model (LLM) Mistral outperformed Llama and rivals the performance of GPT-4 Turbo in a real-world application that assessed the completeness of clinical histories accompanying radiology imaging orders from the emergency department, researchers have reported.
A team led by David Larson, MD, from the Stanford University School of Medicine AI Development and Evaluation (AIDE) Lab locally adapted three LLMs -- open-source Llama 2-7B (Meta) and Mistral-7B (Mistral AI) and closed-source GPT-4 Turbo -- for the task of extracting structured information from notes and clinical histories accompanying imaging orders for CT, MRI, ultrasound, and radiography. The study findings were published February 25 in Radiology.
Digging into a total of 365,097 previously extracted clinical histories, the team arrived at 50,186 histories for evaluating each model's performance in extracting five key elements of the clinical history:
- Past medical history
- Nature of the symptoms, description of injury, or cause for clinical concern (what)
- Focal site of pain or abnormality, if applicable (where)
- Duration of symptoms or time of injury (when)
- Clinical prompt, such as the ordering provider’s concern, working differential diagnosis, or main clinical question (clinical concern)
The team also compared the models' results to those of two radiologists and used the best-performing model to assess the completeness of the large set of clinical history entries (with completeness defined as the presence of the 5 key elements, above).
"Incomplete clinical histories are a common frustration among radiologists, and previous improvement efforts have relied on tedious manual analysis," wrote Larson and colleagues. "The clinical history accompanying an imaging order has long been recognized as important for helping the radiologist understand the clinical context needed to provide an accurate and relevant diagnosis."
Toward quality improvement and filling the gaps, the team used 944 nonduplicated entries (284 CT, 21 MRI, 129 ultrasound, and 510 x-ray) to create prompts and foster in-context learning for the three models.
In the adaptation phase of the study, Mistral-7B outperformed Llama 2-7B, with a mean overall accuracy of 90% versus 79% and a mean overall BERTScore of 0.95 versus 0.92. Mistral-7B was further refined to a mean overall accuracy of 91% and mean overall BERTScore of 0.96 and then tasked alongside GPT-4 Turbo to run through 48,942 unannotated, deidentified clinical history entries to represent a real-world application.
"Without in-context input-output pairs (in-context learning examples), the model outputs were inconsistent and unstructured, and the model could 'hallucinate' information," the authors said, noting that only models that underwent prompt engineering and in-context learning with 16 in-context input-output pairs were assessed.
Larson's team found the following:
- Both Mistral-7B and GPT-4 Turbo showed substantial overall agreement with radiologists (kappa, 0.73 to 0.77) and adjudicated annotations (mean BERTScore, 0.96 for both models; p = 0.38).
- Mistral-7B rivaled GPT-4 Turbo in performance, demonstrating a weighted overall mean accuracy of 91% versus 92%; p =0.31), despite Mistral-7B being a "substantially smaller" model.
- Using Mistral-7B, 26.2% of unannotated clinical histories were found to contain all five elements, and 40.2% contained the three highest-weight elements (that is, past medical history; the nature of the symptoms, description of injury, or cause for clinical concern; and clinical prompt).
- The greatest agreement between models and radiologists was for the two elements "when" and "clinical concern," suggesting that these elements are less subjective.
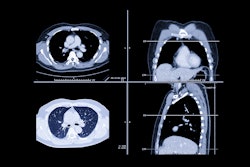
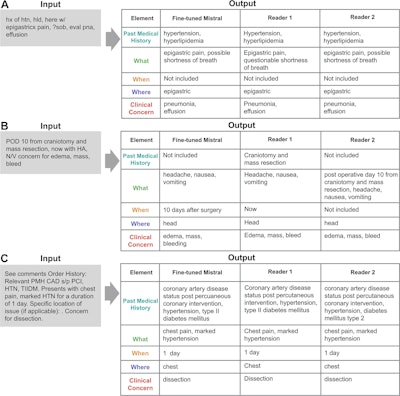 Example input clinical history entries with the outputs generated by the fine-tuned Mistral-7B model and the two radiologists (reader 1 and reader 2). Authors of the paper noted that misspellings in this image are intentional. Graphic and caption courtesy of the RSNA.
Example input clinical history entries with the outputs generated by the fine-tuned Mistral-7B model and the two radiologists (reader 1 and reader 2). Authors of the paper noted that misspellings in this image are intentional. Graphic and caption courtesy of the RSNA.
"These results are promising because smaller models require fewer computing resources, facilitating their deployment," Larson and colleagues wrote. "The fine-tuned Mistral-7B can feasibly automate similar improvement efforts and may improve patient care, as ensuring that clinical histories contain the most relevant information can help radiologists better understand the clinical context of the imaging order."
Additionally, open-source models are unaffected by unpublicized model updates and can be fully deployed locally, avoiding some clinical data privacy concerns with external servers required by proprietary (or closed-source) models, the authors explained. Their next steps include exploring the possibility of not only using the tool to measure and monitor the quality of information shared within Stanford Medicine but also as an educational tool for clinical trainees to help demonstrate "complete" versus "incomplete" clinical histories.
The model code can be found on the Stanford AIDE Lab GitHub repository.
Read the full study here.