Using deep learning with T2-weighted MR imaging is a feasible way to assess patients' body composition, researchers have reported.
The findings suggest an alternative to dual-energy x-ray absorptiometry (DEXA) or CT imaging, which have been commonly used for this purpose but which carry particular limitations, wrote a team led by Johannes Haubold, MD, of the University Hospital Essen in Germany. The results were published February 19 in Investigative Radiology.
"MRI offers excellent soft tissue contrast, making it well-suited for visualizing and quantifying different body compartments and tissues regarding their quality and quantity," the group noted. "T2-weighted MRI sequences, in particular, are essential to detect pathologies in the nervous and musculoskeletal systems and provide a clear distinction between muscle and adipose tissue, allowing for accurate body composition analysis."
Interest in using deep learning for body composition analysis (BCA) has continued to grow, as the technology can offer rapid and automated ways to measure body features like muscle or fat volume for indications ranging from oncology, obesity evaluation, and treatment monitoring to sports medicine and rehabilitation, the investigators wrote. They noted that "especially in diagnostics, body composition analysis applied to radiological image data offers the advantage of opportunistic screening by analyzing image data beyond the primary question, potentially identifying otherwise undetected health issues."
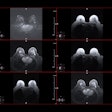
The use of deep learning can make MR imaging for BCA even more efficient by performing automated image segmentation. Haubold and colleagues sought to explore the effect of a deep-learning model for automatic BCA using MR T2-weighted sequences. First, the team generated BCA segmentations of 10 body regions and four body parts by mapping CT segmentations from a body and organ analysis model to synthetic MR images (created using an in-house trained CycleGAN). Thirty synthetic MRI data pairs were used to train two deep-learning segmentation models (nnU-Net V2, 2D, and 3D); the group then used these models to segment 120 real T2-weighted MRI sequences from 120 patients, evaluating the models' performance using Dice scores.
Regarding the 3D ensemble segmentation model used with real MR images, the team found the following Dice scores (with 1 as reference) for various body regions:
- Bone, 0.926
- Muscle, 0.968
- Subcutaneous fat, 0.98
- Nervous system, 0.973
- Thoracic cavity, 0.978
- Abdominal cavity, 0.989
- Mediastinum, 0.92
- Pericardium, 0.945
- Brain, 0.966
- Glands, 0.905
The investigators also found that the 2D ensemble model had the highest Dice scores for all body parts:
- Arms, 0.952
- Head and neck, 0.965
- Legs, 0.978
- Torso 0.99
"The overall average Dice across body parts and body regions ensemble models indicates stable performance across all classes," the team wrote.
What's next? More research, according to the authors.
"While our study demonstrates BCA-MR's promise, further research is needed to explore its full potential in clinical settings," they concluded. "Validation in larger and more diverse patient groups is crucial to ensure generalizability. Additionally, research is needed to explore the utility of BCA-MR in specific clinical scenarios, such as monitoring treatment responses for obesity, sarcopenia, or rehabilitation."
The complete study can be found here.